Google Deepmind has unveiled the third major version of its “AlphaFold” – AlphaFold 3, artificial intelligence model, designed to help scientists design drugs and target disease more effectively.
Structure of Proteins
- Proteins are the polymers of α-amino acids and they are connected to each other by peptide bond or peptide linkage.
- There are twenty amino acids found in the body, which assist in producing thousands of distinct proteins in the body.
- Proteins may have one or more polypeptide chains.
- Each polypeptide in a protein has amino acids linked with each other in a specific sequence and it is this sequence of amino acids that is said to be the primary structure of that protein.
- Any change in this primary structure i.e., the sequence of amino acids creates a different protein.
Functions of Proteins
- Digestion – Digestive enzymes
- Messenger function
- Movement
- Structure and Support – Keratin, a structural protein
- Cellular communication
|
What is AlphaFold?
- An AI tool developed by Google’s DeepMind in 2018 to predict how proteins fold.
- This artificial intelligence technology that helps scientists understand the behavior of the microscopic mechanisms that drive the cells in the human body.
- Solution to Protein Folding Problem: An early version of AlphaFold, released in 2020, successfully solved the “the protein folding problem.”
- Each protein is made up of a string of smaller building blocks called amino acids, which contain all the information to transform proteins — from a single sequence to a folded, functional 3D structure.
Enroll now for UPSC Online Course
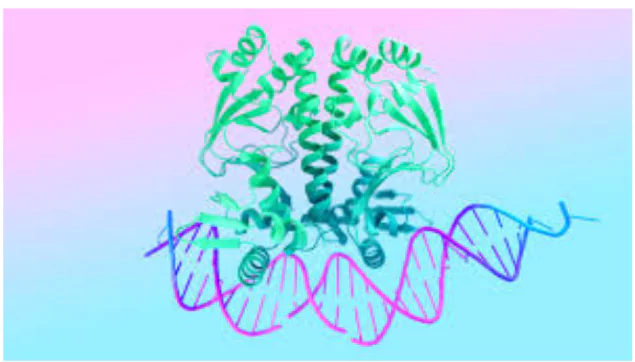
AlphaFold 3
- It’s built on the foundations of AlphaFold 2 & had made a fundamental breakthrough in protein structure prediction in 2020, by predicting the 3D structure of a protein from its amino acid structure.
- It expands beyond proteins to provide accurate predictions for protein interactions with other biomolecules in living cells – such as DNA, RNA, and small molecules.
- Significance: It can predict the behaviour of other microscopic biological mechanisms, including DNA, where the body stores genetic information, and RNA, which transfers information from DNA to proteins.
- It will potentially help to streamline the creation of new drugs and vaccines.
From Noise To Signal
- Original AlphaFold: It was trained on the thousands of sequences and protein structures present in the protein data bank, a giant protein repository where scientists submit experimentally determined protein structures.
- Unlike its predecessors: AlphaFold 3 uses a diffusion model, which is what image-generating software also uses.
-
- The model works by first training on protein structures, adding noise to the data, and then trying to de-noise it.
- This way, the model becomes able to work its way back from a noisy structure to a real protein structure.
- This architecture also helps AlphaFold 3 handle a much larger input dataset.
|
![]() 21 Jun 2024
21 Jun 2024

