Genetic engineering researchers have discovered a next generation genomic design method known as the Bridge Recombinase Mechanism.
- The findings are reported in the form of two papers that were published in the journal Nature characterising the discovery and the working of a ‘bridge’ RNA molecule which can then be reprogrammed as needed, and the structural mechanism behind the discovered recombination ability of these genes.
About Bridge Recombinase Mechanism
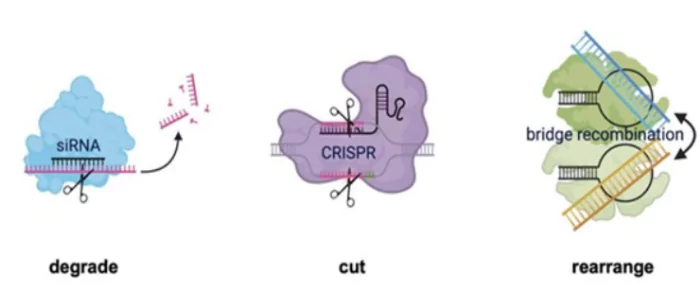
This tool will allow researchers to rearrange, recombine, invert, duplicate, move, and perform other editing operations on very long DNA sequences.
- Existence: This gene editing method exists naturally and has now been discovered, enhances the human ability to edit genomes beyond the capabilities and scope of CRISPR (clustered regularly interspaced short palindromic repeats).
- CRISPR is a technology that can be used to modify the DNA of living organisms.
- It utilises mobile genetic elements or “jumping genes”, which cut and paste themselves into genomes and are present in all forms of life, performing on-the-go DNA manipulation through all living beings.
- Role of Transposons or Jumping Genes: The bits of extra DNA at the ends of jumping genes get joined together and convert the DNA double helix structure into a single-stranded RNA molecule that folds into two loops.
- Binding: This can then bind to two sets of DNA, the donor and the target, with each loop of the element binding independently to the donor segment and target segment.
- Bridging: The target DNA segment is the one that needs to be modified, and the donor segment is the one whose parts will be used to modify the target sequence. Thus, this jumping gene then functions as a bridge that recombines two bits of unconnected DNA.
- Independent Working: The donor loop and the target loop can be programmed independently, offering great flexibility in inserting or recombining sequences to DNA.
- IS110: It is the name of the jumping gene, which stands for Insertion Sequence, and such sequences are found in ample quantities in bacteria (E. coli).
-
- In E. coli bacteria, the bridge RNA had more than 60% insertion efficiency (i.e. ability to introduce a desired gene) and a 94% specificity (ability to target the intended location on the genome).
- They roam around the body, cutting and pasting themselves, repairing DNA and modifying it daily.
- The IS110 bridge recombination system expands the diversity of nucleic-acid-guided systems beyond CRISPR and RNA interference, offering a unified mechanism for the three fundamental DNA rearrangements — insertion, excision and inversion that are required for genome design.
- Structure: The mechanism reports results from cryo-electron microscopy (a technique in which samples are cooled to cryogenic temperatures to determine three-dimensional structures of internal fragments).
-
- The cryogenic temperature range has been defined as from −150 °C to absolute zero or −273 °C.
- By using cryo-electron microscopy to study the IS110 transposons, it is found that it works as a dimer, a complex compound formed by bonding two copies of a simpler compound.
- One copy binds to the target DNA and the other binds to the donor DNA, bridged by the bridge RNA.
- Significance: A Big Boon for Synthetic Biology: The technique can be used to manage, or even treat, a wide variety of genetic diseases: a functional copy of a gene can be replaced in a given genomic location.
-
- The role of the flanking DNA (sequences found on either side of the DNA fragment of interest) present around the recombinase genes of these mobile sequences has been understood for the first time.
- Future Scope: Researchers may also be able to treat chromosomal inversions or deletions, which are currently beyond the reach of any of the editing tools we have.
- It is expected to lead to more advanced gene editing therapeutics and treatments for diseases.
- Limitations: As RNA bridge modifies large sequences of DNA instead of just a tiny segment consisting of a handful of genes, this technique modifying large chunks of DNA sequences increases the risk of unintentional consequences.
- A limitation of the findings is that the studies were performed in vitro or in the lab on bacteria. To apply to any humans, they would need to be tried on animal models, and specifically mammal models, first.
Enroll now for UPSC Online Classes
Other Gene-editing Technologies
- CRISPR-Cas9: A customizable tool that lets scientists cut and insert small pieces of DNA at precise areas along a DNA strand.
- TALE nucleases: Nucleases that cleave unique genomic sequences in living cells can be used for targeted gene editing.
- Zinc-finger nucleases: Targeted to cleave a chosen genomic sequence and provokes cellular repair processes that in turn mediate efficient modification of the targeted location.
- RNA interference (RNAi): Targets RNA molecules to block or activate gene expression.
How does RNA bridge differ from CRISPR, RNAi?
- On Express/Activate: Both CRISPR and RNA Interface (RNAi) work by blocking gene expression or activating it. RNAi does so by targeting RNA molecules, which then do the work of editing genes, while CRISPR edits DNA directly.
- Scope of Change: CRISPR-mediated editing sometimes leaves small bits of nucleotides added/deleted during the repair process. DNA recombination mediated by bridge RNA on the other hand makes a clean cut, making the edit specific and tidy.
- Also, RNA bridge can facilitate the addition, deletion or inversion of DNA sequences of virtually any length.
Check Out UPSC NCERT Textbooks From PW Store
About Transposons or Jumping Genes
These are minimal segments of DNA that have the recombinase enzyme, which binds this DNA to other DNA, along with extra DNA segments at the ends of the genes.
- Discovery: Barbara McClintock found that some genes were able to move around within the genome. These genes were called mobile elements or transposons.
- Between 1948 and 1983, researchers found transposons in an array of life-forms, including bacteriophages, bacteria, plants, worms, fruit flies, mosquitos, mice, and humans. They were nicknamed ‘jumping genes’.
- Working: Prof. McClintock also made another significant observation: depending on where the mobile elements were inserted, they had the ability to reversibly alter gene expression.
- She used corn kernels’ colours as a surrogate to understand hereditary characteristics, and this way figured out transposons moved about in the genome of the maize plant.
- Achievement: She was awarded the Nobel Prize in Physiology or Medicine in 1983 for this work.
- Significance of Transposons:
- Revolutionising the Understanding: The discovery of transposons revolutionised understanding of genetics, in particular their role in enabling nature’s diversity.
- Tools of Evolution: Transposons influence the effects of genes by turning ‘on’ or ‘off’ their expression using a variety of epigenetic mechanisms. They are thus rightly called the tools of evolution, for their ability to rearrange the genome and introduce changes.
- More than 45% of the human genome consists of transposable elements.
- Concern:
- Inactiveness: Transposons also create mutations in genes and lead to diseases. However, most of the transposons have themselves inherited mutations and have become inactive, and thus can not move around within the genome.
- Actions Taken: Over the years, researchers have attempted to resurrect inactive transposons from the genomes of the animal kingdom, hoping that the results will be useful in biomedical applications like genetic correction to cure a disease or for gene therapy.
-
- Example: In 1997, researchers studied the genomes of fish and reconstructed a transposon called ‘sleeping beauty’ at the molecular level.
- Researchers have already discovered several naturally occurring vertebrate transposons and continue to look for more.
About DNA Insertion
- It is a genetic process in which a segment of DNA is added to a different DNA segment, excision is a mechanism in which a damaged DNA segment is removed, and inversion is a method in which a piece of DNA in a chromosome gets reversed.
|
![]() 8 Jul 2024
8 Jul 2024
